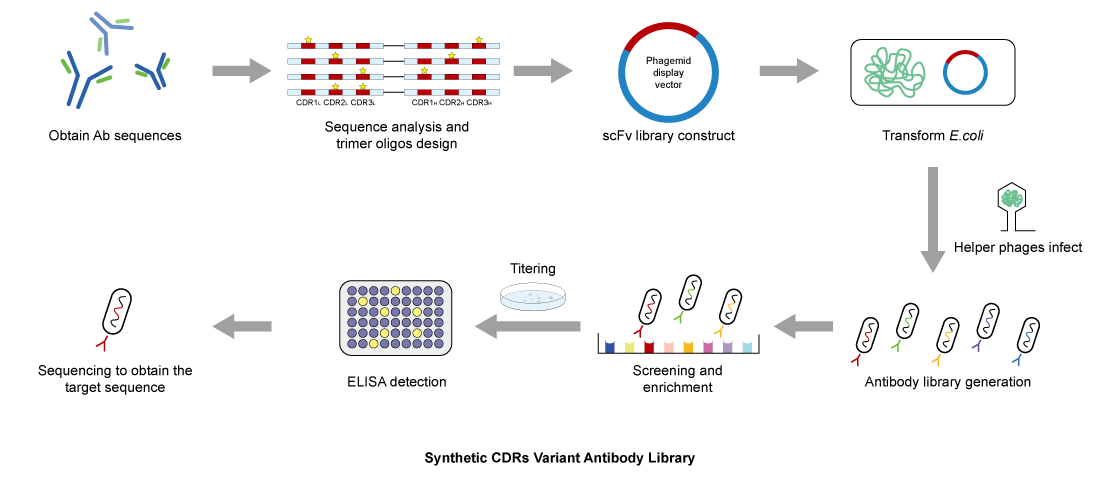
The General Introduction of Synthetic CDR Variant Antibody Libraries
Synthetic antibody libraries are instrumental in designing, constructing, and successfully generating highly functional
antibodies for various applications. These libraries feature optimized framework sequences (a robust scaffold) and controlled CDR diversity, giving them unique advantages over natural antibodies.
Synbio Technologies uses novel Trimer oligonucleotide-directed mutagenesis for codon optimization and synthesis of antibody CDR3 or all CDRs to create variant regions scFv or nanobody libraries. We have successfully delivered hundreds of high-quality, customized variant libraries. The antibody libraries, constructed with precise Trimer primers, provide technical support for antibody affinity maturation, drug target screening, specific antibody discovery, and other fields.
Highlights
-
![]()
- Precisely Targeting CDRs
-
![]()
- Diverse Synthetic Antibody Libraries
-
![]()
- Enhanced Phage Display System
-
![]()
- Efficient & Economical
Service Details
|
Process
|
Details
|
Turnaround Time
|
|
Sequence analysis and trimer oligos design
|
Optimize framework sequence and design the amino acid composition and proportion of the CDRs.
|
1-2 weeks
|
|
Vector construction
|
Sequence assembly and cloning in the antibody expression vectors.
|
2-3 weeks
|
|
Transform E.coli and Library construction
|
Transform the constructs into TG1 competent cells, then prepare the plasmid and identify the capacity and accuracy of the library.
|
2-3 weeks
|
Workflow
Deliverables
-
Synthesized DNA fragments
-
Synthesized libraries cloned into vectors
-
Variable fragment phagemid library
FAQs

Antibody library synthesis aims to construct large-scale libraries through synthetic DNA methods. These libraries contain up to billions of antibody sequences that cover a wide range of antigen binding sites, and provide abundant resources for the screening and discovery of specific antibody drugs.
In order to ensure the diversity and quality of any antibody library, there are some key strategies to follow:
• Design Strategy: adopt a reasonable antibody sequence design strategy to cover maximal possible antigen binding sites.
• High-quality Gene Synthesis: use advanced gene synthesis technology to ensure the accuracy of the synthesized DNA fragments.
• Efficient Cloning Approach: optimize cloning and transformation conditions to improve efficiency and library capacity.
• Strict Library Quality Control: conduct rigorous quality control testing on the constructed library, including assessments of library capacity and diversity.
Some applications of the antibody library synthesis service are:
• Drug Development: used in screening and discovering antibody drugs with therapeutic potential.
• Diagnostic Reagent Development: used in preparing diagnostic antibodies with high sensitivity and specificity to diseases.
• Basic Research: provides abundant antibody reagents for basic research in the field of life sciences.
Get in Touch with Us
Related Services
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries CRISPR Libraries
CRISPR Libraries Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard CRISPR Libraries
Standard CRISPR Libraries Standard CRISPR Plasmid
Standard CRISPR Plasmid ProXpress
ProXpress Protein Products
Protein Products